Predicting the (yet) Unknown: The CAPRI Challenge
Eisenstein, M.1, Ben-Zeev, E.2, Berchanski,
A.2, Heifetz, A.2 and Shapira, B.2
1 Department of Chemical Services, Weizmann Institute of Science
2 Department of Biological Chemistry, Weizmann Institute of Science
Prediction of the structures of protein-protein complexes is an important and fast developing field in the current structural-genome era. In the past few years we witness a vast increase in the number of publications in this field, which describe a variety of prediction (docking) methods.
All docking methods use the structures of the individual molecules in the complex and try to answer the question "How do they combine to form a complex?" Commonly, the prediction methods are tested first on ‘bound structures’, attempting to re-assemble protein-protein complexes using the structures of the complexed molecules. Next, the structures of the uncomplexed molecules are used in ‘unbound docking’, which is a more realistic test of the given docking method. Notably, all the docking methods report much better results for ‘bound docking’ compared to ‘unbound docking’. This is due to a variety of reasons. First, most of the protein-protein docking methods treat the molecules as rigid bodies whereas the structures of the uncomplexed molecules differ from their structure in the complex. In addition, most docking methods include only a part of the interaction energy terms, and often represent them in an approximate form. Nevertheless, success does not elude us and in many cases, good predictions are obtained even in ‘unbound docking’. But how do we compare the different docking methods? Each group has its own selection of complexes used for the development and testing of their method; some complexes are easier to predict and others are harder, and each group may or may not publish the results of only some of their prediction attempts.
The CAPRI (Critical Assessment of PRediction of
Interactions) challenge is a blind docking test that provides a common basis
for comparison of different docking methods. The participants are given the
structures of the individual molecules and are requested to send their
predicted structures of the complexes by a given date, after which the
experimental structures are made public. An independent group of assessors
tests and compares all the predictions. In the first CAPRI challenge there
were 3 prediction targets and 16 predicting groups, which submitted 0-10
solutions per target.
We were the only group that
submitted an acceptable prediction for each of the 3 targets.
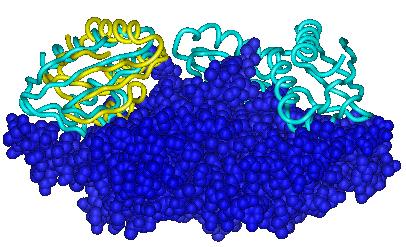
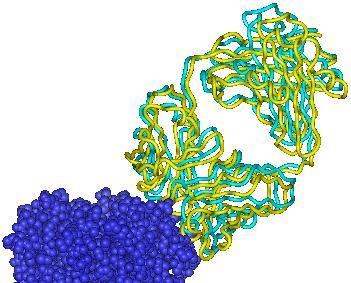
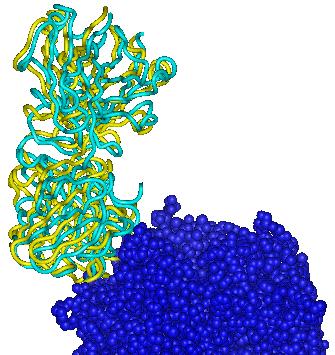
Each figure below presents a superposition of the predicted position of the ligand (yellow ribbon) onto the experimental structure (cyan ribbon).

|

|

|
| Target 1 | Target 2 | Target 3 |