Formal Modeling, Simulation and Analysis of C. elegans Development
Kam, N.1,2, Marelly, R.1, Kugler, H.1, Cohen, I.R.2, Harel, D.1,
Pnueli, A.1, Stern, M.J.3 and Albert Hubbard,
E.J.4
1 Department of Computer Science and Applied Mathematics, Weizmann
Institute of Science
2 Department of Immunology, Weizmann Institute of Science
3 Department of Genetics, Yale University School of Medicine,
New Haven, CT, USA
4 Department of Biology, New York University, New York, NY, USA
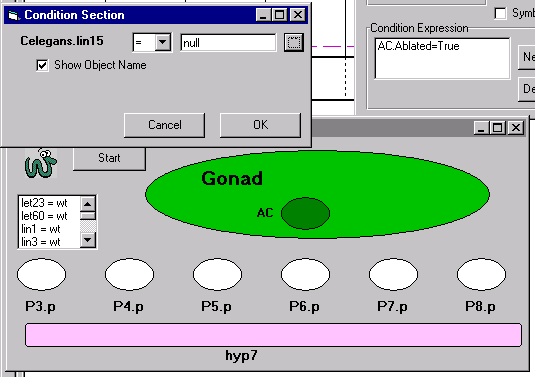
The field of developmental genetics is entering a new phase, in which the synthesis of information from many sources will be necessary to gain a deeper understanding of how various tissues, cells, biochemical interactions and genetic networks collaborate to form a functional organism. The purpose of this project is to model and rigorously simulate and analyze a particular biological system — the C. elegans egg-laying system — using languages, methodologies and tools developed by computer scientists for the reliable development of highly reactive computerized systems. The model will incorporate existing anatomical, genetic and biochemical data pertaining to the development and function of (i) the gonad, (ii) the vulva, (iii) the uterine and vulval musculature, and (iv) the hermaphrodite specific neurons (HSNs). We concentrate on an object-oriented approach using the visual language of statecharts for specifying behavior, and tools such as Rhapsody for model execution and analysis. In previous work, we have successfully applied this language and tool to the biological phenomenon of T cell activation. The T cell activation model served as a feasibility test and integrated phenomena associated with cell-cycle control, cell fate, cell behavior and location. We are now in the midst of a far more ambitious effort, involving more complex biological phenomena that will incorporate additional aspects of development, including cell fate acquisition, cell migration, axon guidance, and apoptosis. In principle, our model will eventually handle virtually all aspects of development, ultimately allowing our results to be extended to and used by the entire C. elegans community, and will apply to other systems too.
As a first stage, we aim at formalizing the existing genetic, biochemical and anatomical data from the biological literature into a set of live sequence charts (LSCs). These LSCs capture the behavior of the system in terms of inter-object behavior, describing the interaction between objects as scenarios. LSCs enable the user to distinguish between scenarios that can occur in the system, scenarios that must occur in the system, and ones that are forbidden ("anti-scenarios"). Within this aim is the development of a graphical user interface (GUI) for the C. elegans. This part of the project will use a recently developed system called the Play-Engine, which enables the user to input the behavioral information in a user-friendly way, and to execute it too. Thus, non-computer scientists can enter biological data in ways in which they are accustomed to representing their system. This will become a critical point regarding one of the future plans of this project: enabling the entire C. elegans community to ‘play-in’ experimental data into a behavioral database of LSCs.

For additional information, demonstration and references see:
http://www.wisdom.weizmann.ac.il/~kam/CelegansModel/CelegansModel.htm